Epigenetic Chromatin Regulation
Genetics, cell biology and development

Small RNA-mediated transcriptional gene silencing is a fundamental process that has been observed in many different eukaryotes including fungi, plants, flies, worms and mammals. One of its main tasks is to neutralize the activity of transposable elements (TEs), which otherwise destabilize our genome and potentially cause diseases. Small RNAs use their base complementarities to TEs to specifically silence them. However, how cells ensure TE-silencing without disturbing expressions of normal genes is not fully understood.
The ciliated protozoan Tetrahymena identifies TE-derived sequences by a germline-some genome comparison mechanism using small RNAs during programmed DNA elimination, which provides fascinating examples of epigenetic genome regulations and important insights into the interaction between TEs and host genomes. Because programmed DNA elimination can be synchronously induced in laboratory in a large scale, it serves as a useful laboratory model for genetically and biochemically investigating small RNA-mediated chromatin regulation.
Using this tiny-hairy eukaryotic model, we aim to understand: how cells accumulate small RNAs specifically from TE-related sequences; how cells use those small RNAs to identify TE-related sequences; and how a small RNA pathway establishes silent chromatin environment (heterochromatin) on TE-related sequences.
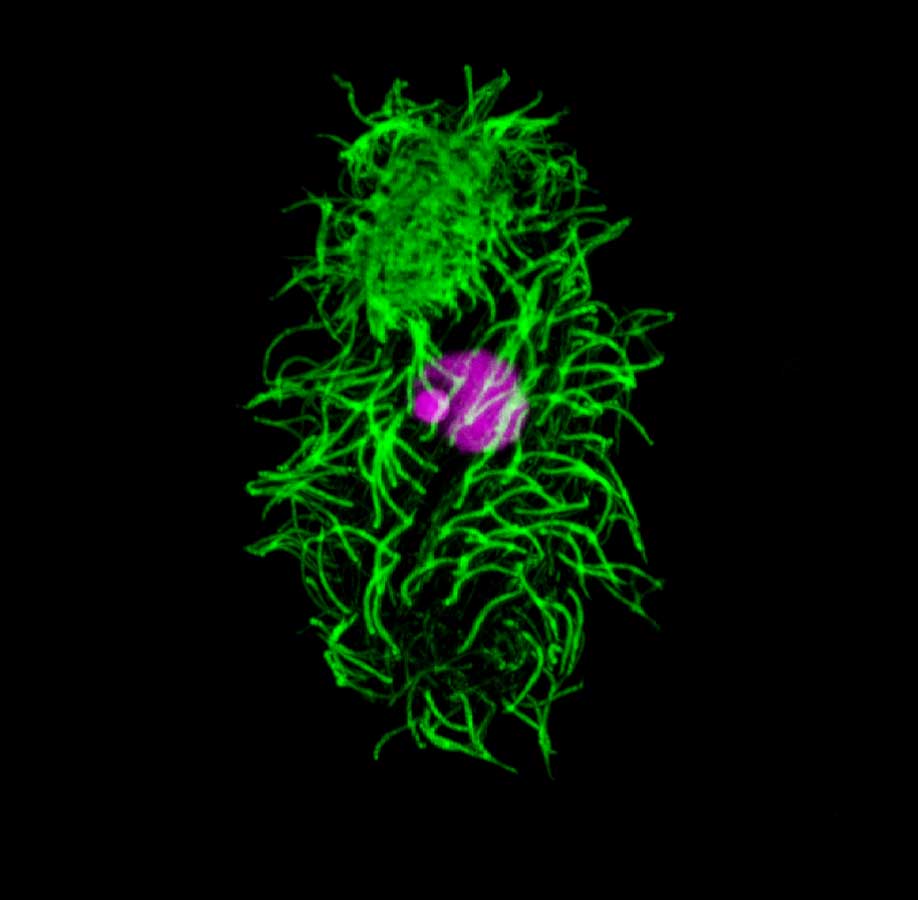
Tetrahymena thermophila. Tetrahymena is a unicellular eukaryote. Tetrahymena has many cilia on its cell surface (green = anti-alpha tubulin staining). Tetrahymena has two different nuclei (stained purple), the smaller germline micronucleus (MIC) and the larger somatic macronucleus (MAC).
Publications of the team
Programmed DNA elimination: New metazoan models.
Mochizuki K
PIWI-Directed DNA Elimination for Tetrahymena Genetics.
Shehzada S, Mochizuki K
Arrested crossover precursor structures form stable homologous bonds in a Tetrahymena meiotic mutant.
Tian M, Mochizuki K, Loidl J
Genomes: Programmed DNA Elimination in a Parasitic Nematode.
Mochizuki K
Non-coding RNA Transcription in Tetrahymena Meiotic Nuclei Requires Dedicated Mediator Complex-Associated Proteins.
Tian M, Mochizuki K, Loidl J
Diversification of small RNA amplification mechanisms for targeting transposon-related sequences in ciliates.
Mutazono M, Noto T, Mochizuki K.
Small RNA-Mediated trans-Nuclear and trans-Element Communications in Tetrahymena DNA Elimination.
Noto T, Mochizuki K
Whats, hows and whys of programmed DNA elimination in Tetrahymena.
Noto T, Mochizuki K
A Zip3-like protein plays a role in crossover formation in the SC-less meiosis of the protist Tetrahymena.
Shodhan A, Kataoka K, Mochizuki K, Novatchkova M, Loidl J
Negative Regulators of an RNAi-Heterochromatin Positive Feedback Loop Safeguard Somatic Genome Integrity in Tetrahymena
Suhren JH, Noto T, Kataoka K, Gao S, Liu Y, Mochizuki K.
Heterochromatin aggregation during DNA elimination in Tetrahymena is facilitated by a prion-like protein.
Kataoka K, Mochizuki K
Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome.
Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, Russ C, Zeng Q, Fan L, Levin JZ, Shea T, Young SK, Hegarty R, Daza R, Gujja S, Wortman JR, Birren BW, Nusbaum C, Thomas J, Carey CM, Pritham EJ, Feschotte C, Noto T, Mochizuki K, Papazyan R, Taverna SD, Dear PH, Cassidy-Hanley DM, Xiong J, Miao W, Orias E, Coyne RS
Phosphorylation of an HP1-like protein is a prerequisite for heterochromatin body formation in Tetrahymena DNA elimination.
Kataoka K, Noto T, Mochizuki K
Phosphorylation of an HP1-like Protein Regulates Heterochromatin Body Assembly for DNA Elimination.
Kataoka K, Mochizuki K
Targeted Gene Disruption by Ectopic Induction of DNA Elimination in Tetrahymena.
Hayashi A, Mochizuki K
Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination.
Noto T, Kataoka K, Suhren JH, Hayashi A, Woolcock KJ, Gorovsky MA, Mochizuki K
A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms.
Woehrer SL, Aronica L, Suhren JH, Busch CJ, Noto T, Mochizuki K
The taming of the shrew: Regulation of a catalytically active domesticated transposase.
Vogt A, Mochizuki K
A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena.
Vogt A, Mochizuki K
Epigenetics of ciliates.
Chalker DL, Meyer E, Mochizuki K
Analysis of Piwi-loaded small RNAs in Tetrahymena.
Noto T, Kurth HM, Mochizuki K
Transposon domestication versus mutualism in ciliate genome rearrangements.
Vogt A, Goldman AD, Mochizuki K, Landweber LF
Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena.
Schoeberl UE, Kurth HM, Noto T, Mochizuki K
Developmentally programmed, RNA-directed genome rearrangement in Tetrahymena.
Mochizuki K
DNA rearrangements directed by non-coding RNAs in ciliates.
Mochizuki K
Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism.
Kataoka K, Mochizuki K
Keeping the soma free of transposons: programmed DNA elimination in ciliates.
Schoeberl UE, Mochizuki K
RNA-directed epigenetic regulation of DNA rearrangements.
Mochizuki K
Modules for C-terminal epitope tagging of Tetrahymena genes.
Kataoka K, Schoeberl UE, Mochizuki K
MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena.
Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J
A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila.
Cheng CY, Vogt A, Mochizuki K, Yao MC
The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus.
Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K
Two GW repeat proteins interact with Tetrahymena thermophila argonaute and promote genome rearrangement.
Bednenko J, Noto T, DeSouza LV, Siu KW, Pearlman RE, Mochizuki K, Gorovsky MA
Non-coding RNA: a bridge between small RNA and DNA.
Kurth HM, Mochizuki K
2'-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena.
Kurth HM, Mochizuki K
High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene.
Mochizuki K
Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena.
Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K
DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena.
Mochizuki K, Novatchkova M, Loidl J
A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase.
Mochizuki K, Gorovsky MA
RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs.
Mochizuki K, Gorovsky MA
Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement.
Mochizuki K, Gorovsky MA
Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena.
Liu Y, Mochizuki K, Gorovsky MA
Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena.
Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA
Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra.
Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T
Expression and evolutionary conservation of nanos-related genes in Hydra.
Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T
Thèses et hdr
Regulations of small RNA turnover during programmed DNA removal in Tetrahymena 28/10/2022
Defended by Salman Shehzada on october 28th 2022 under the direction of Kazufumi Mochizuki
Characterisation of proteins required for RNA-mediated heterochromatin formation 04/12/2020
Defended by Elliot Geraud the 04/12/2020