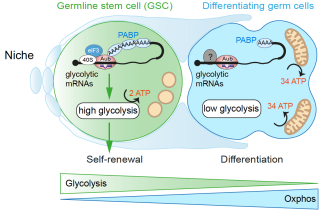
piRNAs are regulators of metabolic reprogramming in stem cells
Rojas-Ríos P, Chartier A, Enjolras C, Cremaschi J, Garret C, Boughlita A, Ramat A, Simonelig M
Genetics, cell biology and development
Gene regulation at the post-transcriptional level is of crucial importance in a wide number of biological processes. The main focus of our lab is to understand mRNA regulation in developmental and pathological processes. We are using the Drosophila model, an outstanding model for in vivo studies.
We have recently identified the role of a specific class of small non-coding RNAs, called Piwi-interacting RNAs (piRNAs) in the regulation of maternal mRNAs during early embryogenesis. piRNAs have been discovered in the last decade in germ cells of animal species. They are loaded into specific Argonaute proteins, the PIWI proteins and their main function is to repress transposable elements in the germline. piRNAs are mostly produced from transposable element sequences. In the last years, we pioneered the discovery of a new function of piRNAs and PIWI proteins in gene regulation. Specifically, piRNAs produced from transposable elements target and repress maternal mRNAs that encode proteins essential for embryonic patterning. These data allowed us to propose the new concept of gene regulation by piRNAs. Furthermore, we revealed a developmental function of transposable elements, a component of the genome whose function has not been clarified. Further studies in other organisms have demonstrated the evolutionary conservation of piRNA function in cellular mRNA regulation. Thus, gene regulation by piRNAs should have a major impact in many biological processes, including diseases.
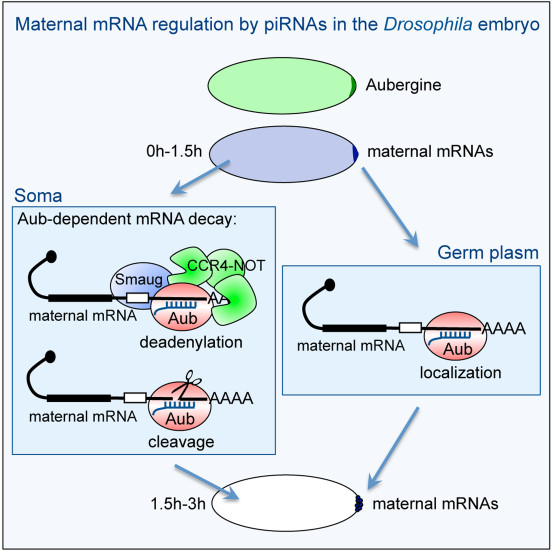 Figure : Scheme of the molecular mechanisms of maternal mRNA regulation by piRNAs and the PIWI protein Aubergine, in the Drosophila embryo (Barckmann et al. Cell Reports 2015; Dufourt et al. Nature Communications 2017).
Figure : Scheme of the molecular mechanisms of maternal mRNA regulation by piRNAs and the PIWI protein Aubergine, in the Drosophila embryo (Barckmann et al. Cell Reports 2015; Dufourt et al. Nature Communications 2017).
Our projects aim at understanding the molecular mechanisms of gene regulation by the piRNA pathway during developmental processes: embryonic development; germline development; stem cell biology; as well as in pathologies.
We have started the series of biennial EMBO Workshops on piRNAs and PIWI proteins in Montpellier (http://events.embo.org/16-piRNA/; http://meetings.embo.org/event/18-pirna).
With the aim of deciphering the importance of mRNA regulation in a pathological context, we have developed a Drosophila model of oculopharyngeal muscular dystrophy (OPMD), a rare genetic disease due to mutations in Poly(A) binding protein nuclear I (PABPN1). PABPN1 has essential functions in nuclear polyadenylation and alternative poly(A) site choice (APA). OPMD results from short alanine expansions at the N terminus of PABPN1. It is characterized by the progressive weakening of specific muscles. Alanine expanded PABPN1 forms nuclear aggregates. OPMD is therefore a proteinopathy, as are Alzheimer and Huntington diseases. Our Drosophila model reproduces the characteristics of the disease. We are using this model to understand the molecular mechanisms underlying disease progression and to identify potential therapies.

Rojas-Ríos P, Chartier A, Enjolras C, Cremaschi J, Garret C, Boughlita A, Ramat A, Simonelig M
Anne Ramat, Ali Haidar, Céline Garret, Martine Simonelig
Naït-Saïdi R, Chartier A, Abgueguen E, Guédat P, Simonelig M
Wang X, Ramat A, Simonelig M, Liu MF
Aude Guénolé, Fabien Velilla, Aymeric Chartier, April Rich, Anne-Ruxandra Carvunis, Claude Sardet, Martine Simonelig & Bijan Sobhian
+
Laboratory of Molecular Virology

Ribot C, Soler C, Chartier A, Al Hayek S, Naït-Saîdi R, Barbezier N, Coux O, Simonelig M
Anne Ramat, Martine Simonelig
Anne Ramat, Maria-Rosa Garcia-Silva, Camille Jahan, Rima Naït-Saïdi, Jérémy Dufourt, Céline Garret, Aymeric Chartier, Julie Cremaschi, Vipul Patel, Mathilde Decourcelle, Amandine Bastide, François Juge & Martine Simonelig
Malerba A, Roth F, Harish P, Dhiab J, Lu-Nguyen N, Cappellari O, Jarmin S, Mahoudeau A, Ythier V, Lainé J, Negroni E, Abgueguen E, Simonelig M, Guedat P, Mouly V, Butler-Browne G, Voisset C, Dickson G, Trollet C
Rojas-Ríos P, Simonelig M
Coll O, Guitart T, Villalba A, Papin C, Simonelig M, Gebauer F
Barckmann B, Dufourt J, Simonelig M
Dufourt J, Bontonou G, Chartier A, Jahan C, Meunier AC, Pierson S, Harrison PF, Papin C, Beilharz TH, Simonelig M
Rojas-Ríos P, Chartier A, Pierson S, Simonelig M
Götze M, Dufourt J, Ihling C, Rammelt C, Pierson S, Sambrani N, Temme C, Sinz A, Simonelig M, Wahle E.
Chartier A, Joly W, Simonelig M.
Rojas-Rios, P., Chartier, A., Pierson, S., Severac, D., Dantec, C., Busseau, I., Simonelig, M.
Barckmann,B., Pierson,S., Dufourt, J., Papin, C., Armenise, C., Port, F., Grentzinger, T., Chambeyron, S., Baronian, G., Desvignes, JP., Curk, T., Simonelig, M.
Chartier A, Klein P, Pierson S, Barbezier N, Gidaro T, Casas F, Carberry S, Dowling P, Maynadier L, Bellec M, Oloko M, Jardel C, Moritz B, Dickson G, Mouly V, Ohlendieck K, Butler-Browne G, Trollet C, Simonelig M.
Simonelig M
Temme, C., Simonelig, M., Wahle E.
Joly, W,, Chartier,A., Rojas-Rios, P., Busseau,I., Simonelig, M.
Barckmann B, Simonelig M.
Chartier A, Simonelig M.
Simonelig, M.
Papin C, Simonelig M.
Simonelig, M
Anvar SY, 't Hoen PA, Venema A, van der Sluijs B, van Engelen B, Snoeck M, Vissing J, Trollet C, Dickson G, Chartier A, Simonelig M, van Ommen GJ, van der Maarel SM, Raz V.
Barbezier N, Chartier A, Bidet Y, Buttstedt A, Voisset C, Galons H, Blondel M, Schwarz E, Simonelig M.
Rouget, C., Papin, C., Boureux, A., Meunier, A.C., Franco, B., Robine, N., Lai, E.C., Pelisson, A. & Simonelig, M.
Temme, C., Zhang, L., Kremmer, E., Ihling, C., Chartier, A., Sinz, A., Simonelig, M., Wahle, E.
Chartier, A., Raz, V., Sterrenburg, E., Verrips, CT., van der Maarel, SM., Simonelig, M
Benoit, P., Papin, C., Kwak, JE., Wickens, M., Simonelig, M.
Chicoine, J., Benoit, P., Paliouras, M., Gamberi, C., Simonelig, M. and Lasko, P.
Zaessinger S, Busseau I, Simonelig M.
Chartier A, Benoit B, Simonelig M.
Benoit, B., Mitou, G., Chartier, A., Temme, C., Zaessinger, S., Wahle, E., Busseau I., Simonelig, M.
Temme, C., Zaessinger, S., Meyer, S., Simonelig, M., Wahle, E.
Juge, F., Zaessinger, S., Temme, C., Wahle, E. and Simonelig, M.
Benoit, B., Juge, F., Iral, F., Audibert, A. and Simonelig, M.
Juge, F., Audibert, A., Benoit, B. and Simonelig, M.
Benoit, B., Nemeth, A., Aulner, N., Kuhn, U., Simonelig, M., Wahle, E. and Bourbon, H.M.
Audibert, A. and Simonelig, M.
Audibert, A. and Simonelig, M.
Audibert, A., Juge, F. and Simonelig, M.
Defended by Rima Nait-Saidi on 23-11-2022
Defended by Camille Jahan on 29-03-2019
