Evaluation of microRNA variant maturation prior to genome edition.
Busseau I, Mockly S, Houbron É, Somaï H, Seitz H
Genetics, cell biology and development
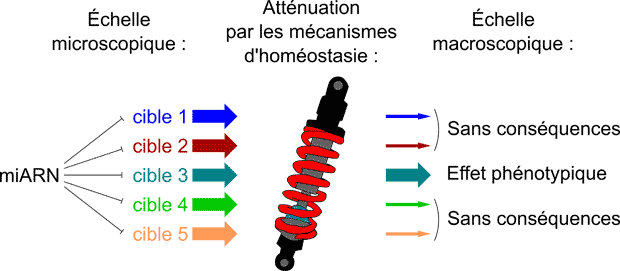
MicroRNAs ("miRNAs") are small post-transcriptional regulators. The function of these small RNAs in animals has been well characterized at a molecular level, but their role is less well known at the macroscopic scale: how could miRNAs have any biological function if they repress most of their targets less than 2-fold (while inter-individual gene expression fluctuation typically exceeds 2-fold, and is buffered by homeostatic mechanisms)?
According to the current dogma, each miRNA regulates tens or hundreds of targets, yet several observations suggest miRNAs have a much weaker impact on animal biology. Our recent work also suggests that both experimental and computational miRNA target identification methods are heavily contaminated with false positives: these false positives may be truly repressed by miRNAs at the molecular scale, but such a small repressive effect fails to translate into a macroscopic phenotype for most genes.
Our work thus suggests that the biological role of miRNAs has been largely over-estimated. We are currently exploring practical consequences of this new theoretical framework, measuring the contribution of individual miRNA/target interactions to global in vivo phenotypes.
More generally, we are proposing a new vision of gene regulation: a regulatory target is not simply a gene that is affected by a regulatory pathway; it is a gene that is affected enough by the pathway – the extent of a measured regulation needs to be confronted to the robustness of biological systems to fluctuations.
Busseau I, Mockly S, Houbron É, Somaï H, Seitz H
Houbron É, Mockly S, Rafasse S, Gros N, Muriaux D, Seitz H
Mockly S, Seitz H
Sophie Mockly, Élisabeth Houbron, Hervé Seitz
Hebras J, Marty V, Personnaz J, Mercier P, Krogh N, Nielsen H, Aguirrebengoa M, Seitz H, Pradere JP, Guiard BP, Cavaille J
Patricia Richard, Shuang Feng, Yueh-Lin Tsai, Wencheng Li, Paola Rinchetti, Ubayed Muhith, Juan Irizarry-Cole, Katharine Stolz, Lionel A Sanz, Stella Hartono, Mainul Hoque, Saba Tadesse, Hervé Seitz, Francesco Lotti, Michio Hirano, Frédéric Chédin, Bin Tian, James L Manley
Canzler S, Schor J, Busch W, Schubert K, Rolle-Kampczyk UE, Seitz H, Kamp H, von Bergen M, Buesen R, Hackermüller J
+
Systemic impact of small regulatory RNAs
Canzler S, Schor J, Busch W, Schubert K, Rolle-Kampczyk UE, Seitz H, Kamp H, von Bergen M, Buesen R, Hackermüller J
+
Systemic impact of small regulatory RNAs
Mockly S, Seitz H
Amar L, Seitz H
Seitz H
Pinzón N, Bertrand S, Subirana L, Busseau I, Escrivá H, Seitz H
Marlétaz F, Firbas PN, Maeso I, Tena JJ, Bogdanovic O, Perry M, Wyatt CDR, de la Calle-Mustienes E, Bertrand S, Burguera D, Acemel RD, van Heeringen SJ, Naranjo S, Herrera-Ubeda C, Skvortsova K, Jimenez-Gancedo S, Aldea D, Marquez Y, Buono L, Kozmikova I, Permanyer J, Louis A, Albuixech-Crespo B, Le Petillon Y, Leon A, Subirana L, Balwierz PJ, Duckett PE, Farahani E, Aury JM, Mangenot S, Wincker P, Albalat R, Benito-Gutiérrez È, Cañestro C, Castro F, D'Aniello S, Ferrier DEK, Huang S, Laudet V, Marais GAB, Pontarotti P, Schubert M, Seitz H, Somorjai I, Takahashi T, Mirabeau O, Xu A, Yu JK, Carninci P, Martinez-Morales JR, Crollius HR, Kozmik Z, Weirauch MT, Garcia-Fernàndez J, Lister R, Lenhard B, Holland PWH, Escriva H, Gómez-Skarmeta JL, Irimia M
Buesen R, Chorley BN, da Silva Lima B, Daston G, Deferme L, Ebbels T, Gant TW, Goetz A, Greally J, Gribaldo L, Hackermüller J, Hubesch B, Jennen D, Johnson K, Kanno J, Kauffmann HM, Laffont M, McMullen P, Meehan R, Pemberton M, Perdichizzi S, Piersma AH, Sauer UG, Schmidt K, Seitz H, Sumida K, Tollefsen KE, Tong W, Tralau T, van Ravenzwaay B, Weber RJM, Worth A, Yauk C, Poole A
Rodríguez-Martínez, M., Pinzón, N., Ghommidh, C., Beyne, E., Seitz, H., Cayrou, C., Méchali, M.
+
Replication and Genome Dynamics
Seitz H
Pinzon, N., Li, B., Martinez, L., Sergeeva, A., Presumey, J., Apparailly, F., Seitz, H
Eckenfelder A, Ségéral E, Pinzón N, Ulveling D, Amadori C, Charpentier M, Nidelet S, Concordet JP, Zagury JF, Paillart JC, Berlioz-Torrent C, Seitz H, Emiliani S, Gallois-Montbrun S.
Aigner A, Buesen R, Gant T, Gooderham N, Greim H, Hackermüller J, Hubesch B, Laffont M, Marczylo E, Meister G, Petrick JS, Rasoulpour RJ, Sauer UG, Schmidt K, Seitz H, Slack F, Sukata T, van der Vies SM, Verhaert J, Witwer KW, Poole A
Hoffmann S, Clauss S, Berger IM, Weiß B, Montalbano A, Röth R, Bucher M, Klier I, Wakili R, Seitz H, Schulze-Bahr E, Katus HA, Flachsbart F, Nebel A, Guenther SP, Bagaev E, Rottbauer W, Kääb S, Just S, Rappold GA.
Royo H, Seitz H, ElInati E, Peters AH, Stadler MB, Turner JM
Tarver JE, Cormier A, Pinzón N, Taylor RS, Carré W, Strittmatter M, Seitz H, Coelho SM, Cock JM
Chambeyron, S., Seitz, H.
+
Non-coding RNA, epigenetics and genome stability
Moran, Y., Fredman, D., Praher, D., Li Z. L., Meng Wee,L., Rentzsch, F., Zamore,P.D., Technau, U., Seitz H.
Sergeeva, A., Restrepo, N.P., and Seitz, H.
Seitz H, Tushir JS, Zamore PD
Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y
Defended by Sophie Mockly on 08/12/2021 under the supervision of Hervé Seitz
Website of the event : https://internat21.sciencesconf.org/.
Website of the event : https://internat.sciencesconf.org.
Content of the introductory lectures given during the first half-day of the summer school (in French)(introduction on RNAi, on microRNAs, and on the biochemistry of the RISC complex; exercises): freely downloadable here.
| Collaborator | Aim of the collaboration | Common publications |
|---|---|---|
| Dr. Yukihide TOMARI (université de Tõkyõ) Web site |
Analysis of structural determinants of microRNA biogenesis | Kawamata et al. (2009) Pubmed Tsutsumi et al. (2011) Pubmed |
| Dr. Ulrich TECHNAU (université de Vienne) Web site |
Functional analysis of small regulatory RNAs in Nematostella vectensis | Moran et al. (2014) Pubmed |
| Dr. Fabian RENTZSCH (SARS Center) Web site |
Functional analysis of small regulatory RNAs in Nematostella vectensis | Moran et al. (2014) Pubmed |
| Prof. Phillip D. ZAMORE (école de médecine de l'université du Massachusetts) Web site |
Functional analysis of small regulatory RNAs in Nematostella vectensis | Moran et al. (2014) Pubmed |
| Dr. James TURNER (Crick Institute, Londres) Web site |
Analysis of the expression of microRNAs subjected to an epigenetic control in mammals | Royo et al. (2015) Pubmed |
| Dr. Denis TAGU (INRA Rennes) Web site |
Characterization of small regulatory RNAs in pea aphid | |
| Dr. Florence APPARAILLY (IRMB, Montpellier) Web site |
Measurement of inter-individual fluctuation in gene expression in neutrophils | Pinzón et al. (2017) Pubmed |
| Dr. Marcel MÉCHALI (IGH, Montpellier) Web site |
Identification of replication origins in early Nematode development and epigenetic characterization | Rodríguez-Martínez et al. (2017) Pubmed |
| Dr. Mark COCK (UMR 8227, Roscoff) Web site |
MicroRNA identification in the brown alga Ectocarpus silicosus | Tarver et al. (2015) Pubmed |
| Prof. Gudrun RAPPOLD (UniversitätKlinikum, Heidelberg) Web site |
Identification of a microRNA binding site specific to patients with atrial fibrillation | Hoffmann et al. (2016) Pubmed |
| Dr. Hector ESCRIVA (Observatoire océanologique de Banyuls) Web site |
Characterization of small RNAs in the cephalochordate Branchiostoma lanceolatum |
Marlétaz et al. (2018) Pubmed Pinzón et al. (2019) Pubmed |
| Dr. Sarah GALLOIS-MONTBRUN (Institut Cochin, Paris) Web site |
Identification of Ago binding sites in HIV-1 viral RNA | Eckenfelder et al. (2017) Pubmed |
| Dr. James Manley (Columbia university, New York, États-Unis) Web site |
Characterization of molecular consequences of SETX repression in human cells | Richard et al. (2020) Pubmed |
| Dr. Jérôme Cavaillé (CBI, Toulouse) Web site |
Evaluation of molecular and physiological consequences of the deletion of the SNORD115 RNA |
Hebras et al. (2020) Pubmed |
Scripts and datasets for the preparation of figures and supplementary figures - 56 KB
Scripts and datasets for the preparation of figures and supplementary figures - 7,2 GB
Scripts and datasets for the preparation of figures, supplementary figures and supplementary tables - 636Mo
Scripts and datasets for the preparation of Table 1
Scripts and datasets for the preparation of all 6 figures :
Scripts and data for the generation of figures 2D, 3B and 4A
Conversion script (usage : ./read_FCS.py input.fcs output.txt) : script python.
1. Unix training
2. Statistics in molecular biology
3. R training
Context :
CNRS researchers have to submit every year a yearly activity report, and every 5 years they have to submit a “phased activity report” which is more detailed (as well as a “mid-phase activity report” in the middle of that 5 year-period). Following the merge of regions Midi-Pyrénées and Languedoc-Roussillon, laboratories in Languedoc-Roussillon joined the phase of Midi-Pyrénées laboratories, thus delaying by one year the evaluation which was scheduled for 2019 (it will take place in 2020).
In addition, laboratories are evaluated every 5 to 6 years by an international committee, which is assembled by HCERES (formerly known as AERES), which will then issue an evaluation report (in English).
Activity reports and evaluation reports for our team:
2022 mid-phase activity report of Hervé Seitz (December 2019 – June 2022) (in French)
2020 evaluation report of our team by HCERES (in English)
2020 phased activitiy report of Hervé Seitz (January 2014 – December 2019) (in French)
2016 mid-phase activity report of Hervé Seitz (January 2014 – September 2016) (in French)
2014 evaluation report of our team by AERES (in English)
2014 phased activity report of Hervé Seitz (January 2009 – December 2013) (in French)
« La technologie CRISPR/Cas9 »
(compte-rendu de la conférence donnée par Hervé Seitz au colloque « Quelles limites pour les technosciences en santé ? » à Clermont-Ferrand le 13 mars 2018, et publié dans le n°15 de la Revue du Centre Michel de l'Hospital).
Parallèle entre l'informatique et la génétique
(édition du génome, débuggage, matérialité de l'information, ...) (interview donnée dans le cadre des Tic-Talks du LIG).
Les statistiques en science expérimentale. Principe, limitations, erreurs courantes, illustrées par les rumeurs pseudo-scientifiques sur la Covid-19
(visio-conférence donnée le 13 mars 2021 sur invitation du cercle zététique du Languedoc-Roussillon ; diaporama cliquable accessible ici).
